Research
"mein Laboratorium ist die Natur [my laboratory is Nature]" —
(Christian) Ernst Stahl
Synopsis
Our lab is interested in understanding the molecular basis for how mutualisms and symbioses evolve and the ecological forces that shape the formation of de novo symbioses and microbial consortia. We prefer to tackle these issues using two approaches: (1) a synthetic, forward/bottom-up approach in which we try to discover new microbial partnerships and build microbial ecosystems, and (2) a more top-down, combined culturing + sequencing/omics-based approach. If culture-independent, metagenomic sequencing is at one end of the proverbial "candle" of understanding microbial ecology and "synthetic ecology" is at the other end, we try to burn the candle at both ends.
A broad interest of ours is to leverage both genomic and phenotypic data of sequenced organisms to better understand (and experimentally test) the capacity for novel species interactions, particularly in new environments. An ongoing mission of the lab is to distill 'rules' for how microbes interact to form stable communities that perform defined functions. We care about both the mechanisms that enable microbes to play well together upon first encounter, as well as to stay together for the long-haul as they change and co-evolve.
We have a number of ongoing projects in the lab, which are briefly described below.
Exploring the Capacity of Fungi to Interact with Photoautotrophic Microbes
Of the estimated number of fungi that are believed exist on this planet (conservatively over 1.5 million species), less than 10% have been identified to date despite fungal diversity efforts over the past decade. Where are they all? Originally supported by a multi-institution NSF GoLife grant,
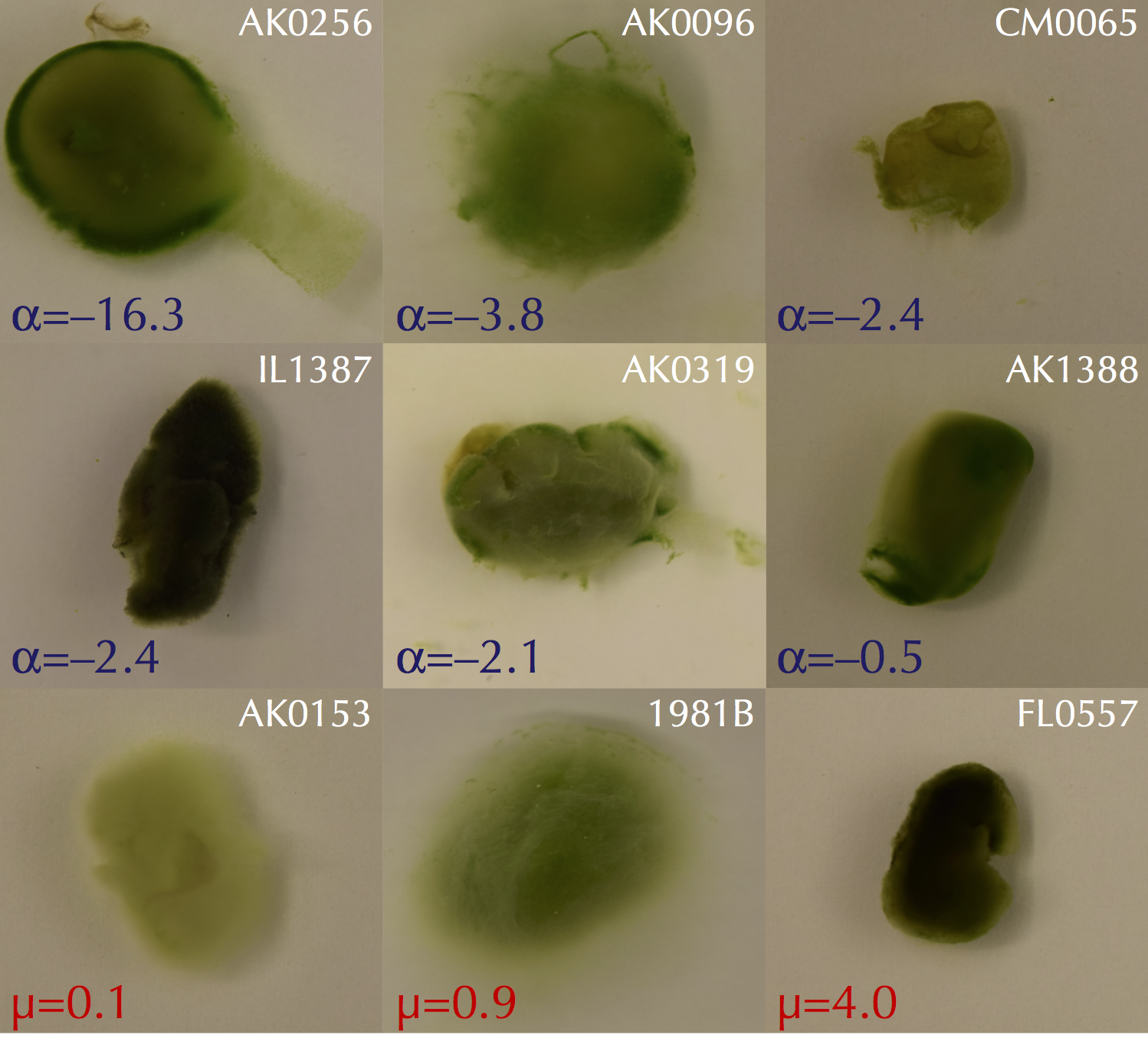
we (with our fantastic collaborators) believe part of the answer may be 'in association with photoautotrophs' – i.e., plants, algae, and cyanobacteria. We have participated in thrilling field sampling trips around the globe in search for new species of fungi and algae/cyanobacteria and complement our field studies with co-culturing and have noticed that the sort of fungal-algal sticking that Erik
reported a decade ago can also be seen with other fungi in a species-specific and nutrient-dependent manner. We are working to understand these "Franken-lichen" associations, and what they can tell us about extant fungal-algal/cyanobacterial associations found in nature. Stay tuned for more updates!
Current Team: Eli, Prabin, Qijie
Experimental Evolution of a Model Microbial Mutualism
A prototypical mutualistic system that we study in the lab involves two 'workhorse organisms' of eukaryotic biology: the budding yeast
Saccharomyces cerevisiae and the unicellular green alga
Chlamydomonas reinhardtii (affectionately dubbed the "green yeast"). Following up on work done by Erik to establish this system, we are pursuing nutristat co-evolution experiments to study how this model mutualism adapts as a function of input nutrient concentrations (carbon and nitrogen), strength of dependency (obligate vs. facultative mutualism), historical contingency (prior evolutionary history), and gene flow. We seek to relate these studies to
John Thompson's Geographic Mosaic Theory of Coevolution, and to test some of the hypotheses posed by Thompson's Theory (and recast) for our microbial ecosystem.
Current Team: Prabin, Destiney, Yang, Christian
Understanding Secondary Metabolite Production and the Chemical Ecology of Marine Polycultures
In collaboration with Mike Lomas at the
NCMA/Bigelow Laboratory of Ocean Science and colleagues at
Ole Miss' National Center for Natural Products Research, we are exploring the potential for marine polycultures in the NCMA collection to produce secondary metabolites (SMs) of therapeutic value. For positive bioassay 'hits,' we try to elucidate the molecular basis for SM production and regulation, as well as the relevance of these SMs to the native ecology of the species contained within the polyculture. Our first lucky attempt led us to identify a suite of related, potent anti-malarial and anti-tumor SM compounds from one marine polyculture. We have identified the gene cluster responsible for at least one SM and are trying to study how this suite of compounds might be relevant in the native ecosystem from which this polyculture was derived. We also have a couple of budding collaborations to investigate polycultures dominated by marine fungi, which has generally been neglected in the mycological mainstream. We hope to have good news soon on all these fronts.
Current Team: Matthew, Emerson, Andrew
Green Microbiome of Sloths
We're sure you've heard of sloths, given their recent meme-ish popularity in movies (
Zootopia) and commercials (
Geico). But did you know that many sloths are "green" because algae/cyanobacteria grow on their hair? In fact, sloths host an array of microbes (including fungi) within their fur, along with moths, mites, and other insects. In collaboration with
The Sloth Institute in Costa Rica, we are working to better understand and characterize the 'green' microbiome of sloths, its potential role in sloth health, and its functions in the sloth fur ecosystem.
Current Team: Matthew, Destiney, Qijie
Water and Milk Kefir

|
We are studying two traditional fermented beverages, water and milk kefir, with the goal of developing them into model systems for testing fundamental principles in microbial ecology and synthetic ecology. Water kefir is made with sugar water while milk kefir is a yogurt-like drink made with milk; both traditionally use "grains" – symbiotic aggregates – to ferment the medium (similar to a SCOBY used to make Kombucha Tea). We are interested in understanding the microbial similarities and differences underlying these two beverages, and to delineate what is common among different heirloom cultures that may have been propagated independently for generations around the world. Each culture typically yields distinct bouquets/flavors and we are interested in integrating microbial, genetic, metabolic, and sensory layers of description to form a holistic understanding of these communities. Ultimately, we'd like to study the microbial dynamics of these "aquatic" ecosystems, and be able to manipulate/engineer specific ecosystem functions.
Current Team: Cas, Danh, Dow, Nicole, Clark, Evan, Kate, Prabin
Biological Soil Crusts
We are collaborating with the labs of
Nicole Pietrasiak and
Jason Stajich to better understand fascinating (but understudied) biological soil crust (BSC) microbial communities in which algae and/or cyanobacteria are keystone species. BSCs serve as ecosystem pioneers in dryland/desert contexts: they are the first entities to colonize barren dry land and pave the way for succession by higher plants and animals. With the help of a
DOE FICUS grant and scientists at the Joint Genome Institute (JGI) and Environmental Molecular Science Laboratory (EMSL), we seek to understand how these BSCs are organized and function using a combination of omics and imaging technologies available uniquely at the JGI/EMSL. BSCs have a neat feature that makes them experimental tractable: they remain in "suspended animation" when dry but become reanimated when wet. Just add water when you're ready!
Current Team: Qijie, Andrew
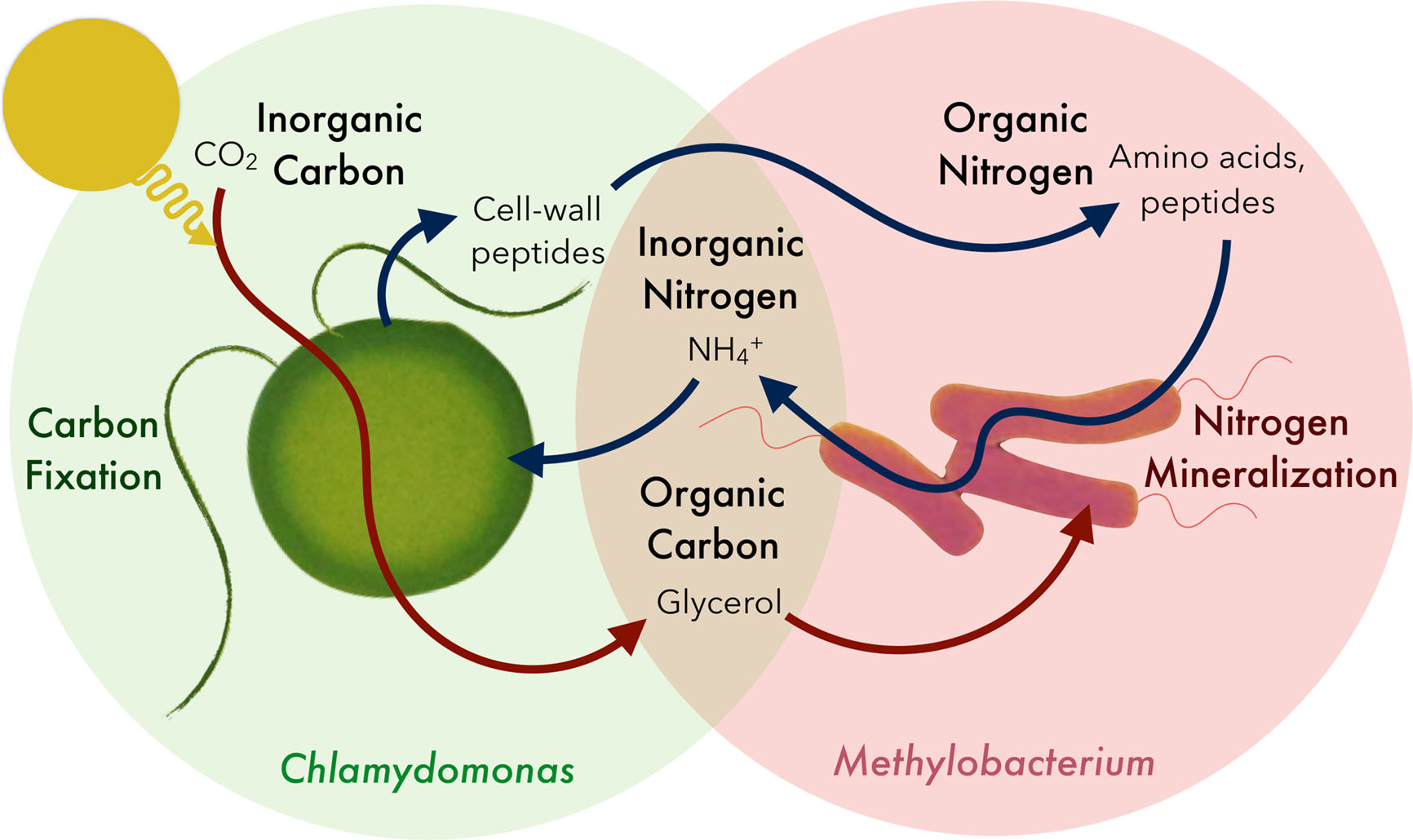
Methylobacterium-Chlamydomonas Mutualism
In collaboration with
Aurora Galván and Emilio Fernández's lab in Córdoba Spain, we have been investigating a serendipitous discovery made by graduate student Victoria Calatrava, who visited our lab in summer of 2016 to kick things off on this project. (You can find a paper describing parts 1 and 2 of this work in the
publications page; a part 3 manuscript is in preparation for submission soon!) Based on complementary nitrogen metabolic abilities, many
Methylobacteria spp. can form mutualisms with the model green alga,
Chlamydomonas reinhardtii. Methylobacteria are interesting in being able to use methanol as a carbon source and are one of the most, if not the most dominant bacterium found on plant leaves, where they thrive on the methanol released from the stomata of plants. And they're pink! Methylobacteria can stimulate seed germination and influence plant development, so we find it intriguing and are following our noses that methylobacterial associations with a 'lower plant' -- i.e., Chlamydomonas -- may yield basic insights into methylobacteria-plant biology. This project is an example of a new microbial partnership that was "created" by chance discovery. Both methylobacteria and
Chlamydomonas reinhardtii reside in soil (and rice fields!), so it's possible that what we observe may recapitulate what occurs in nature.
Current Team: Noah
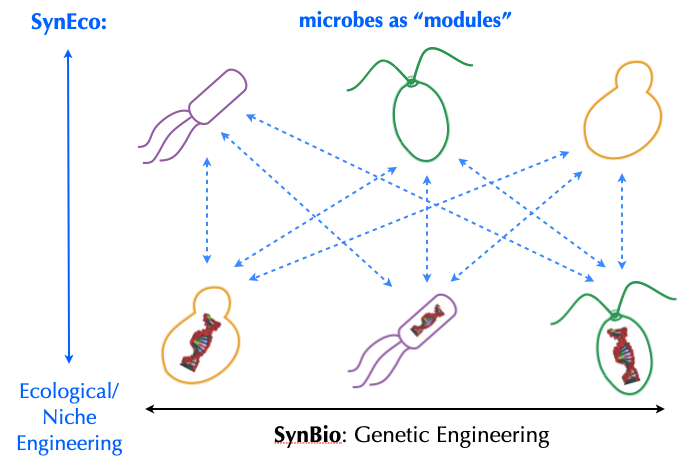
Synthetic Microbial Communities
We're going back to our roots in synthetic ecology with a new effort to assemble and analyze synthetic communities de novo. We are particularly interested in developing methods to disentangle "interactions" within these communities via culture-based and omics-based approaches. We're in quiet mode at the moment as we lay the groundwork, but hope to have progress to report on within a year. Erik recently gave a
short talk at a Symbiosis and Artificial Life workshop at ALIFE2020 in which some of the motivations for this work are articulated.
Current Team: Destiney, Qijie
Tea Plant Endo- and Epi-Microbiome
In collaboration with
Cam Muir, proprietor of Big Island Tea, we have initiated an exploratory project to understand the ecological and microbial drivers of tea quality. This effort was spawned from an experiential learning Study USA course that Erik has led to Hawaii for UM students. We are focusing on characterizing the endo- and epi-phytic microbes of the tea plant
Camelia sinensis on Cam's farm, vis-a-vis forest diversity and insect damage. This is early work on a very complex system, but we believe we will be able to make some fundamental and practical insights relevant to plant-microbe interactions, impacts on plant leaf chemistry, and microbial biodiversity.
Current Team: Noah, Qijie, Christian
Computational Stuff
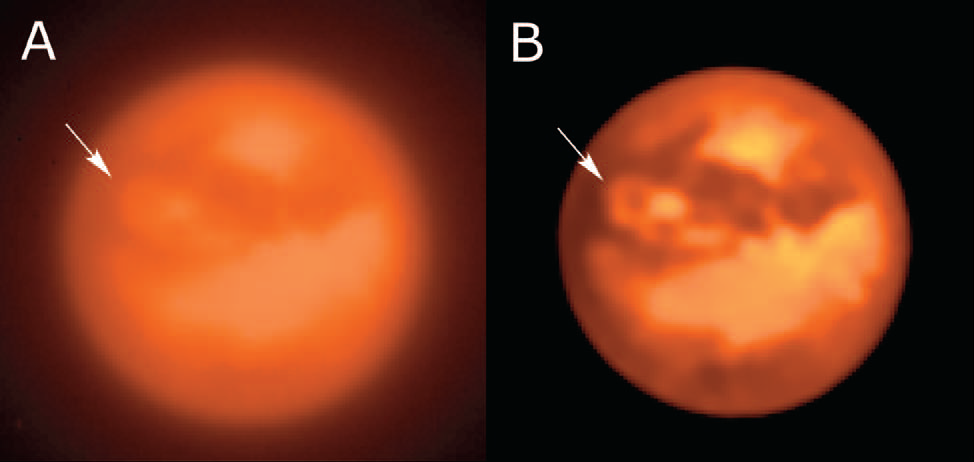
• Even after years of developmental dormancy, the Adaptive Image Deconvolution Algorithm that Erik wrote,
AIDA, is still competitive with the current state-of-the-art. Collaborators,
Franck Marchis and Clemente Chalumeau at SETI have done a good job keeping the ship afloat and have been busy using AIDA for their astronomical work. But with recent local talent and interest from a couple of computer science students, we're finally trying to update and re-tool AIDA to be more efficient and user-friendly, especially for biologists or others who may not be imaging/optics aficionados.
• Erik has a few modeling projects that complement the experimental projects described above. One of these involves establishing an agent-based modeling framework to explore different ecosystem scenarios for synthetic systems similar to the yeast-Chlamydomonas or methylobacterium-Chlamydomonas mutualisms highlighted above. A related project involves developing a simulation platform to interpret and better understand a new experimental technique we're currently developing to query microbial community interactions.
We hope to report more on all of these in the not too distant future!